Translate this page into:
Chromosome microarray: An advanced genetics diagnostics tool for helping patients of recurrent pregnancy loss in modern Bharat: A pilot study
Address for correspondence: Dr. Fauzdar, Head, Clinical Genomics, Redcliffe Labs, H-55, Sector-63, Noida, Gautam Budh Nagar, Uttar Pradesh 201301, India. E-mail: dr.ashishfauzdar@redcliffelabs.com
-
Received: ,
Accepted: ,
How to cite this article: Fauzdar D A. Chromosome microarray: An advanced genetics diagnostics tool for helping patients of recurrent pregnancy loss in modern Bharat: A pilot study. Fertil Sci Res 2023;10:112-6.
Abstract
Purpose:
The aim of this study was to determine the efficacy and diagnostic power of chromosome microarray (CMA) in products of conception (POC) samples from patient from North India patient population.
Methods:
The data analysis included 100 consecutive POC samples received at our laboratory for CMA analysis over a period of 06 months. The POC samples were referred from women aged between 24-42 years ascertained for recurrent pregnancy loss and/or spontaneous abortion (80%), structural abnormalities in ultrasound, absent nasal bone, cardia defect, increased nuchal translucency. All the samples were evaluated by a whole-genome single-nucleotide polymorphism (SNP)-based chromosome microarray.
Results:
The successful results were obtained in 92%, fresh tissue samples with clinically significant abnormalities identified in 18% of specimens that could be the possible reason for the pregnancy loss. The whole chromosome aneuploidies were the most common chromosome abnormalities including trisomy for chr15 (2), chr16 (3), chr18 (3), chr21 (2), chr22 (2), monosomy for chrX (4) and triploidy (2). The 8% of the sample failed the quality matrix and did not yield any results due poor DNA integrity due to degraded sample quality and positive for maternal cell contamination (mcc).
Conclusion:
The small pilot study confirmed that single nucleotide polymorphism (SNP) based CMA is a robust platform, with successful results obtained in >92% of cases. As compared to analysis of POC specimens by karyotyping that fails in 50-60% of cases. CMA can identify whole chromosomal aneuploidy, polyploidy along with maternal cell contamination, thus maximizing sensitivity and decreasing false-negative results. The SNP based CMA platform has shown tremendous potential in understanding the etiology of fetal loss and help in evaluating the recurrence risk for chromosome aneuploidy and assists in determining appropriate management for future family planning in recurrent pregnancy loss patients.
INTRODUCTION
Spontaneous loss of pregnancy is one of the most common and dreaded complication. It has been estimated that approximately 70% of all-human conceptions fail to achieve viability. Approximately 15 to 20% of clinically recognized pregnancies end up as pregnancy loss with approximately 5% of them experiencing two consecutive miscarriages. The exact prevalence of recurrent pregnancy loss (RPL) is difficult to estimate, but many studies state that RPL affects 1 to 2% of women worldwide. However, the burden of recurrent miscarriages in India is higher as compared to that reported in literature worldwide, i.e., around 7.4%.[1] The RPL is defined as a loss of two or more failed pregnancies in the first trimester. At present, there are many studies that have reported various etiologies of RPL in women, but the most common established cause includes antiphospholipid syndrome, hormonal or metabolic disorders, uterine anatomic abnormalities, infections, sperm quality, lifestyle-related disorders, and genetic or chromosome abnormalities. There are recommendations and guidelines from various national and international professional bodies based on the current evidence from the published studies for the evaluation and management of RPL patients [Table 1]. But at present diagnosis and management of RPL can be done only in 50% of patients while the remaining half remains undiagnosed under the idiopathic category with no established diagnosis. The current investigations and interventions recommended by standard guidelines are based on the evidence of moderate, low, or very low quality. Therefore, treatment and interventions are possible only in about half of the cases, with 50% of the cases reported as idiopathic or with no definitive diagnosis established. The counseling process also becomes complex in such patient families with no identifiable cause or definitive diagnosis observed for pregnancy loss.
| Professional | Current Recommendations | Evidence |
|---|---|---|
| ESHRE | Genetic analysis of the pregnancy tissue is done with array-based comparative genomic hybridization (array-CGH) or microarray and it's been recommended based on a reduction in maternal cell contamination. | Strong |
| Genetic analysis of pregnancy tissue though is not routinely recommended in RPL, but it could be performed for explanatory purposes. | Conditional | |
| Parental karyotyping at present not routinely recommended in couple with RPL, but it could be carried out after individual risk assessment. | Conditional | |
| ASRM | Parents should undergo peripheral blood karyotyping for the detection of balanced structural chromosome abnormalities. | Strong |
| Karyotype analysis of product of conception (POC) may be useful in the setting of ongoing therapy of RPL, but there is possibility of maternal tissue contamination in the specimen. | Conditional | |
| ACOG/SMFM | In cases of intrauterine fetal demise or still birth, further cytogenetic analysis is desired. Chromosomal microarray analysis on the fetal tissue (i.e., amniotic fluid or products of conception) is recommended in the evaluation with increased likelihood of obtaining results and improved detection of causative abnormalities. | Recommended |
| FOGSI | Complete genomic hybridization (CGH) microarray is the preferred test as it identifies smaller defects such as microdeletions, single gene defects, and point mutations including aneuploidy. Has least maternal cell contamination and does not require viable, sterile tissue. Fluorescence in-situ hybridization (FISH) test in low resource setting for Ch 3 13, 16, 18, 21, 22, X, and Y | Recommended |
ACOG = The American College of Obstetricians and Gynecologists, ASRM = American Society of Reproductive Medicine, ESHRE = European Society of Human Reproduction and Embryology, FOGSI = The Federation of Obstetric and Gynaecological Societies of India, RPL = recurrent pregnancy loss, SMFM = Society of Maternal Fetal Medicine
TECHNOLOGY ADVANCES
We present here the data of 100 consecutive product of conception specimens analyzed with advanced genetic technology of chromosomes as recommended by professional bodies over a period of 6 months. The specimens were referred to the national referral laboratory with clinical indications including spontaneous abortion, anembryonic pregnancy, early pregnancy loss, first trimester losses, and RPLs to rule out genetic or chromosome abnormality in the patient specimen. The aim of this study was to determine the efficacy and diagnostic power of chromosome microarray (CMA) in products of conception (POC) samples in Indian patient population. The majority of specimens were obtained from women with RPL and/or spontaneous abortion in 80 (80%), structural abnormalities in ultrasound, absent nasal bone, cardiac defect, and increased nuchal translucency. In order to identify chromosomal abnormalities in POC specimen is first examined under a dissecting microscope to remove placental decidua as it is most likely of maternal origin.
All the patient specimens were evaluated by a whole-genome single-nucleotide polymorphism (SNP)-based CMA, a commercially available platform that includes both SNP and copy markers from all 23 pairs of chromosomes. The results obtained through CMA analysis were expressed as copy number variants (CNVs), which is defined as duplicated or deleted segments of DNA of at least 1000 base pair (kb) in size with a difference from representative reference genome. The patient results were reported either as pathogenic or nonpathogenic with or without clinically significant pathogenic variants identified or variants of uncertain significance (vous) based on the current supporting studies and reported cases in the literature. The successful results were obtained in 92%; fresh tissue samples with clinically significant pathogenic abnormalities were identified in 18% of the specimens explaining the possible reason for pregnancy loss. The whole-chromosome aneuploidies were the most common chromosome (chr) abnormalities including trisomy for chr15 (2), chr16 (3), chr18 (3), chr21 (2), chr22 (2), monosomy for chrX (4), and triploids (2) [Figure 1]. There were no results in the eight samples that failed the quality matrix due to various reasons including poor DNA yield or concentration in the sample or poor DNA integrity due to degraded sample quality or due to maternal cell contamination (MCC).
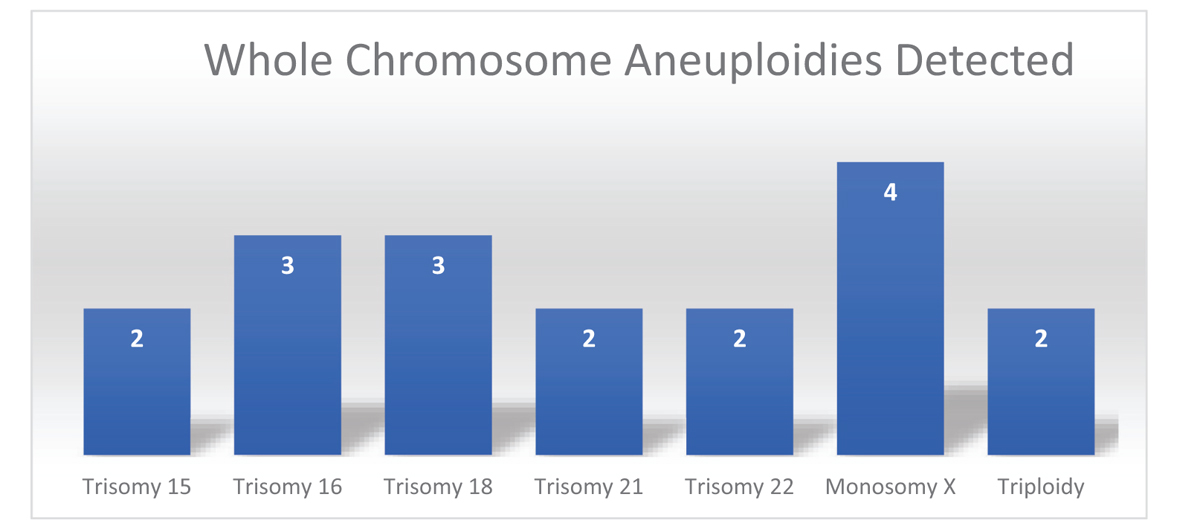
- Bar graph depicting various chromosome abnormalities detected in product of conceptions with chromosome microarray analysis in the study.
Chromosome Microarray (CMA) Technology in Current Obstetrics and Gynecology Practice
Random chromosomal abnormalities are the single most common cause of pregnancy loss of sporadic losses before 10 weeks of gestation and majority of them are due to numerical chromosomal abnormalities (>60%) involving whole chromosomes specifically trisomy, monosomy, and polyploidy as reported from western population. It can be further summarized that pregnancy loss due to chromosome abnormality could be further subdivided into the following three subcategories: (a) numerical chromosomal abnormalities, i.e., trisomy, monosomy (60–80%); (b) structural chromosomal abnormalities including deletion, duplication, inversion, and translocations (2–5%); and (c) polyploidy including triploids or tetraploids (20%) arising due to aberrant fertilization (Fig. 1). The most common structural chromosome abnormality is balanced translocation that could be either Robertsonian translocation (involving same chromosomes) or reciprocal translocation (involving two different chromosomes) and is observed in 2 to 5% of couples with recurrent miscarriages. The most common hypothesis proposed for aneuploidy is random error in segregation or nondisjunction of chromosomes in meiosis I or II during embryogenesis or germ cell development. In recent years, due to advancement of genetic technologies there are various advancement in the genetics methodologies utilized for the examination of embryonic/fetal material (product of conception) and advantages and disadvantages of all technologies are being encapsulated in Table 2.
| Method Characteristics (Technique) | Single Nucleotide Polymorphism- Chromosome Microarray (SNP-CMA) | Karyotype (Conventional Culture Technique) | FISH (Fluorescence ln-situ Hybridization) | QF-PCR (Quantitative Fluorescent- Polymerase Chain Reaction) | NGS (Next-Generation Sequencing) |
|---|---|---|---|---|---|
| Detect | Chromosome abnormalities (aneuploidies, triploidy), unbalanced structural changes (duplication, deletion, amplification) | Changes are chromosome number (aneuploidies, polyploidy)Structural abnormalities (balanced and unbalanced translocation) | Chromosome aneuploidies (13, 18, 21, X, Y, 16, and 22)Diagnosis of submicroscopic chromosome aberrationStructural translocation | Detect aneuploidies for chromosome 13, 18, 21, X, Y, 16, and 22 | Sequencing of large genomic regions, high number of genes with high throughput |
| Samples type | Fresh tissue, FFPE block | Fresh tissue, culture cells | Fresh tissue, uncultured (interphase) cells | Fresh tissue/DNA | Fresh/FFPE |
| Limitation | Cannot detect unbalanced translocation and low level of mosaicism (< 10%) | The requirement of culture of cells (high culture failure rate 10-20%) | Diagnosis specific and limited to probes utilized in kit | Diagnosis within intended use of kit only | Very high sensitivity, excess of information, uninterpretable for diagnosis the genetic cause |
| Culture failures | No | Yes | No | No | No |
| Diagnostic yield | High (50-100 kb) | Low (5-10 MB) | Moderate (100-200 kb) | Moderate | Very high (<50 kb) |
| Maternal cell contamination (MCC) | Yes | No | No | No | No |
| Turnaround time (TAT) for getting results | 7-10 days | 14-21 days | 24-48 hours | <24-48 hours | 3-4 weeks |
| Recommended by guidelines | Yes | Yes | Yes | No | Yes (research) |
| Cost (INR) | 12,000-14,000 | 5000-7000 | 3000-5000 | 3500-6500 | 20,000-25,000 |
FFPE = fresh frozen paraffin embedded, INR = Indian rupee, Kb = kilobases, MB = megabase
CMA is a particularly an attractive technology as it can be performed on a variety of human specimens including product of conception samples, amniotic fluid, peripheral blood, placental villi, fresh frozen paraffin embedded (FFPE) blocks, etc. Since it is performed using extracted cellular DNA from the tissue sample and therefore it significantly improves the detection rate and increased likelihood of obtaining the result for the patient as compared to conventional techniques such as POC karyotype and therefore better management of RPL patient. Another major advantage of DNA-based chromosome analysis through CMA is the improved resolution for detecting chromosomal submicroscopic microdeletions and microduplication and thus increased likelihood for detecting genetic disorders of clinical significance, e.g., Angelman or Prader-Willi syndrome that could not be detected by conventional cytogenetics techniques. Since CMA requires smaller amount of input DNA through automatic workflow and high-throughput equipment therefore ensures a faster turnaround time with multiple samples processing. Notably SNP-based CMA can detect the presence of contaminating maternal cells in product of conception sample therefore drastically reducing the likelihood of a false negative result due to MCC.[2]
CONCLUSION
The SNP-based CMA platform has shown tremendous potential in understanding the etiology of fetal loss and helps in evaluating the recurrence risk for chromosome aneuploidy and assists in the future management of pregnancy. Our study also confirms that SNP-based CMA is a robust platform with successful results obtained in 92% of cases as compared to traditional conventional cytogenetics culture methods that fail in 50 to 60% of the cases. The microarray can identify whole chromosomal aneuploidy, polyploidy along with MCC, thus maximizing sensitivity and decreasing false negatives. CMA will prevent patients from undergoing unnecessary costly investigations once the genetic cause of pregnancy loss is established. In the cases where genetic testing of POC specimen is not available, RPL workup can be started with the genetic evaluation by offering couple karyotype to rule out balanced translocation. Genetic counseling should be offered to couples both pretest and posttest along with informed consent, explaining the advantages and limitations for any advanced genetic investigation performed for the patient.
COMMENTARY
Commentary of “Chromosome microarray: An advanced genetics diagnostics tool for helping patients of recurrent pregnancy loss in modern Bharat: A pilot study”
A very high percentage of human pregnancies are lost during the first trimester. Although the exact reason for this loss remains unknown in most cases, it is believed that many of these are due to chromosomal anomalies. However, when these pregnancy losses become recurrent, with two or more occurrences, it becomes traumatic for the patient and challenging for the clinician to identify the cause, which remains unknown in more than half of such patients.
Karyotyping of the products of conception often does not yield results and is unable to detect microdeletions and microduplications. Therefore, there is a global shift toward more advanced genetic analysis techniques, such as chromosomal microarray for all the chromosomes, which can detect even the single nucleotide polymorphism (SNP) as well as the copy variants.
In this pilot study conducted in India, the authors assessed 100 products of conception to determine chromosomal abnormalities using an SNP-based chromosome microassay. The study aimed to determine the SNP as well as copy number variants. Copy number variants are duplicated or deleted segments of DNA of at least 1000 base pair (Kb) in size, with deviations from the representative reference genome. Results were obtained in 92% of the 100 samples studied, which indicates that this technology may be more effective in assessing products of conception in cases of recurrent pregnancy loss compared to conventionally used methods, which have a failure rate of 40 to 50%. The two limiting factors may be the costs involved and the availability of testing laboratories as well as the efficiency of these laboratories; however, the increasing number of laboratories offering such technologies is promising.
Rajvi Mehta, Consultant, Coopper Surgicals, Trivector Biomed.
REFERENCES
- Standard Treatment Guidelines for Management of Recurrent Spontaneous Abortions. Ministry of Health and Family Welfare, GOI 2017 January
- [Google Scholar]
- Comprehensive genetic analysis of pregnancy loss by chromosomal microarrays: outcomes, benefits, and challenges. Genet Med. 2017;19:83-9.
- [CrossRef] [PubMed] [Google Scholar]






